I was so inspired by a seminar talk from Dr. Jonathan Pritchard yesterday. The talk was about a new conceptual framework “omnigenic model”for dissection of complex traits.
Numerous GWAS showed many genes with small effects compose the architecture of complex traits, and big effect genes only contribute to small proportion of heritability. Where is the “missing heritability” ? They are from widely spread alleles of small effects, which are frequently under detected.
Dr. Pritchard’s group suggested a new framework to solve this question. Simply put, both core genes and peripheral genes contribute to the heritability of a complex trait. The main heritability comes from the trans-eQTL of peripheral genes. The core genes are the key driver of the trait, but they only account for small proportion of heritability. For disease prediction, we need to consider both core and peripheral genes; as for disease mechanism, the core genes, which reside in hub positions in the pathway and influence traits directly, may be more important.
Form evolutionary point of view, big effects genes are more frequently targets for natural selection, because they are easy to be “seen”; while small effects genes are drifting and under neutral selective pressure.
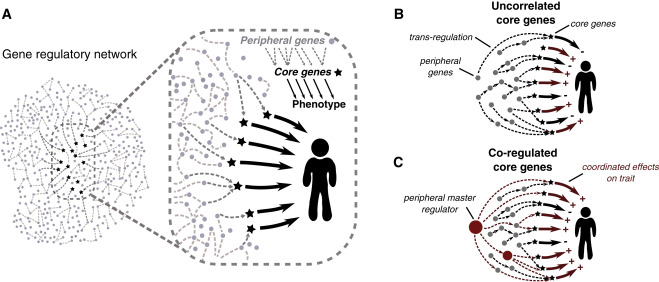
Figure. “Omnigenic model” from https://www.sciencedirect.com/science/article/pii/S0092867419304003
Now, the questions are: how to define core and peripheral genes? Is it possible to set up different threshold for considering these genes when dissecting the architecture of complex traits?